Integrative genomics
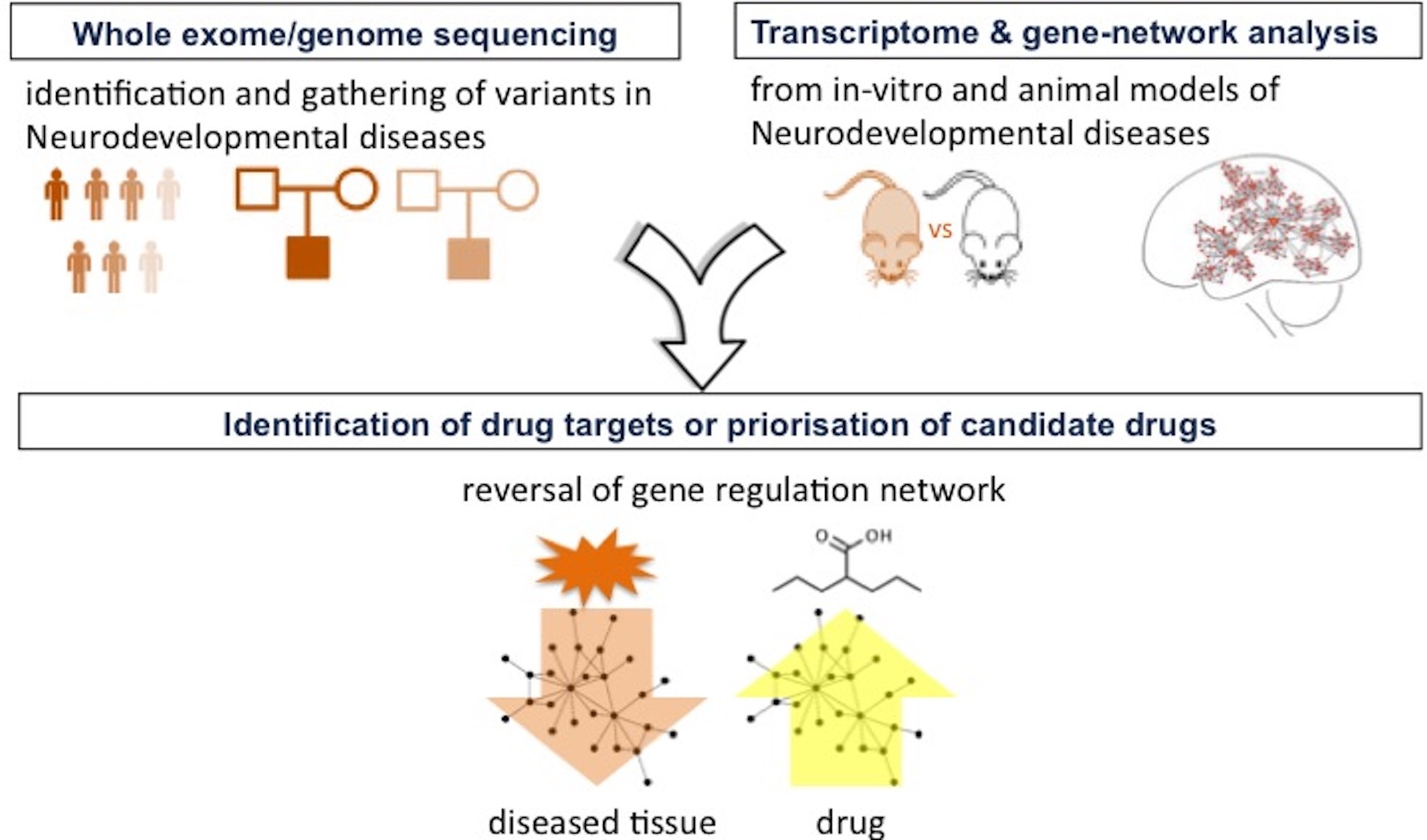
The Integrative Genomics in Neurodevelopment group gathers members interested in normal and diseased brain development, and mainly focuses on statistical and computational biology approaches to integrate -omics data from patients, healthy individuals, animal or in vitro models. Our research applies to neurodevelopmental and neuropsychiatric disorders especially epilepsy, bipolar disorders, and complications of prematurity.
A.D.D
We develop tools to analyze and combine newly generated and existing open-source data. Genome-wide transcriptional profiling, using spatial or single-cell transcriptomics, can help to identify candidate regulators and drivers of the disease under investigation. Our methodological research emphasizes on gene regulatory networks and network analysis to prioritize key regulators that govern developmental and pathological processes. In order to study mood disorders, we use integration of multi-omics data (mRNA-seq, miRNA-seq, methylomes, and GWAS) with phenotype data from patients to identify candidate biomarkers and to help to stratify patient groups. Beyond a better understanding of molecular drivers of neurodevelopment and neuropsychiatric disorders, we aim at providing new candidate targets for drug discovery and repurposing. To this purpose, we currently study the use of machine learning methods to repurpose drugs based on their transcriptional impact.
AI-Assisted Virtual Screening and Structural Assessment of Mutations Identified in Patients (Bruno Villoutreix)
Structure-based virtual screening (SBVS) aims to identify bioactive compounds computationally using the 3D structure of a protein target as input. It involves docking and ranking large numbers of molecules based on predicted binding scores, followed by interactive selection of a short list of molecules for experimental testing. To aid decision-making, ADMET properties are also predicted. Our group specialises in designing protein-protein interaction inhibitors, employing and developing approaches such as docking combined with machine learning to streamline the discovery of hit compounds and drug candidates.
Structural bioinformatics plays a crucial role in investigating the impact of point mutations on protein structure and function. These computational tools attempt to predict how mutations may alter a protein’s 3D structure, stability, and interactions with other molecules. Simulations, docking and energy calculations assess changes in binding affinity, enzymatic activity, or folding, offering insights into molecular mechanisms underlying the health and disease states. When combined with experimental studies, these approaches help bridge the gap between genotype and phenotype.
Contacts
Andrée Delahaye-Duriez
Inserm U1141 – NeuroDiderot
Robert Debré Hospital
48 Boulevard Sérurier
75019 Paris
andree.delahaye@insem.fr
Bruno Villoutreix
Inserm U1141 – NeuroDiderot
Robert Debré Hospital
48 Boulevard Sérurier
75019 Paris
bruno.villoutreix@inserm.fr
Read more

Conference April 16th, 2026, 11:00 AM by Alexandra Benchoua from I-Stem : Human pluripotent stem cell models for high-throughput screening of therapeutic molecules and precision medicine in neurodevelopmental disorders.
Meeting Room 6th floor Bingen Building and Visio https://u-paris.zoom.us/j/84730677369?pwd=AFdhdI6alNWLbXub7sM1DJ33yUIOYN.1 - Summary séminaire A-Benchoua _27-nov-2025 english

Scientific conference Melody Atkins Thursday, April 2nd 2026 (11/00 AM).
Meeting Room 6th floor Bingen Building and Visio https://u-paris.zoom.us/j/84730677369?pwd=AFdhdI6alNWLbXub7sM1DJ33yUIOYN.1

Conference Thursday, January 22, 2026, 11:00 AM by Xavier Leinekugel from the Institut de Neurobiologie de la Méditerranée (INMED), Marseille : Potential consequences of altered maturation of hippocampal perisomatic inhibition on cognitive development and neurodevelopmental disorders
20260122 Xavier LEINEKUGEL english

Conference Thursday, February 12 2026, 2PM (zoom) by Pr Raul Chavez-Valdez from Baltimore : Synaptic Plasticity following Neonatal Hypoxic-Ischemic Brain Injury
20260212 séminaire Raul Chavez english
